The basic principles of the I125 labeling method of antibodies. There are various methods for the iodine labeling of proteins. For example, chemical methods or enzymatic methods are used to iodize protein molecules by oxidation. When the chemical oxidation method is applied, sodium iodide (NaI) meets a strong oxidant, and iodine ions are oxidized into iodine molecules, and the generated free iodine molecules can be halogenated with certain groups. The protein molecules that can undergo halogenation reactions are mainly tyrosine residues, and some histidine residues may also undergo iodination. In the experiment using the Chloramine T (Chloramine T) method, the oxidant (1, 3, 4, 6-tetrachloro-3α, 6α-diphenyl-glucoluril) is soluble in highly volatile organic solvents. After adding the solvent to the test tube, let it volatilize (ie, let the oxidant coat the test tube), and then add Na125I and protein solution to the coated test tube. After the reaction is completed, the mixed solution will be transferred to another tube to terminate the reaction. Reagents and instruments Polyclonal antibody or monoclonal antibody purified by affinity chromatography 0.5 mol / L sodium phosphate buffer, pH 7.5 (see Appendix 1 for the method of preparation) Na125I without carrier 3.7 GBq / ml (100 mCi / ml) NaOH liquid gel filtration column γ-counter 100g / L trichloroacetic acid 70% ethanol glass fiber filter Chloramine T (Chloramine T) reaction * Freshly prepared 0.5 mol / L phosphoric acid containing 2mg / ml chloramine T Sodium buffer (pH 7.5); * Chloramine T reaction termination buffer: 2.4mg / ml partial sulfite, 10mg / ml tyrosine, 10% glycerol, 1g / L Xyene cyanol in PBS.
Operation steps * Note: 125I is harmful to health and requires protective measures. The application of 125I should first be related to isotope knowledge, and under the supervision of relevant departments, according to the application and disposal requirements of radioactive isotopes.
(1) For the chloramine T method, use a 1.5 ml Ependof tube, add 10 μl of antibody and a total volume of 0.5 mol / L sodium phosphate buffer at pH 7.5 to 25 μl; add 500 μCi of Na125I and mix; add 25 μl of 2 mg / ml chloramine T Mix the solution; incubate at room temperature for 1 minute; add 50 μl of chloramine T reaction termination buffer (capture free Na125I with saturated tyrosine); separate the iodinated antibody from the iodinated casein by gel filtration chromatography Acid separation. Put a 1 ml gel filtration chromatography column on the reaction mixture, collect 100 μl of eluent per tube in portions, and discharge the initial component of the iodinated antibody. The components were monitored with a γ-counter; each tube containing the iodinated antibody was collected and combined; the 125I labeled antibody tube was placed in an isotope protection tube and stored at 4 ° C.
(2) Iodogen-coated test tube method Iodogen is dissolved in chloroform at 0.5 μg / ml; 100 μl of Iodogen solution is transferred into a combined glass test tube; the test tube is placed in a fume hood overnight and the chloroform is evaporated at room temperature; 50 μl of antibody (0.2- 1 mg / ml in 0.5 mol / L sodium phosphate buffer at pH 7.5); add 500 μCi Na125I to the antibody-coated Iodogen-coated tube and incubate at room temperature for 2 minutes; transfer the reaction solution in the tube to a 1.5 ml Eppendof tube (In which 50μl Iodogen reaction termination buffer has been added first), mix gently; separate the iodinated antibody from the iodinated tyrosine by gel filtration chromatography (same as the previous complex amine T method); collect and mix the iodinated Each tube of antibody; place the 125I labeled antibody in an isotope protection tube and store at 4 ° C. Experimental quality monitoring: Trichloroacetic acid precipitation method was used to determine the efficiency of antibody labeling: take two pieces of glass fiber filter paper and write a mark with a soft pencil; place the filter paper flat on the hole of the test tube rack so that the center part does not touch the test tube rack; 5μl sample containing about 10000cpm, accurately point to the center of each filter paper, to be dried at room temperature; use flat-tipped tweezers to transfer a filter paper into a test tube, add 2ml 100g / L trichloroacetic acid; filter paper in trichloroacetic acid solution Soak for 10 minutes and pour out the solution; add 2ml of 100g / L trichloroacetic acid and repeat the above operation; add 2ml of 70% ethanol, immerse the filter paper in ethanol and rotate for 10 minutes, then pour out the solution; The radioactivity of the washed filter paper; from the measured amount of 125I bound to the antibody in the spotted filter paper, calculate the total amount of 125I bound to the antibody in the antibody solution. Cpm of washed filter paper cpm of unwashed filter paper × 100 = ratio of bound iodine total sample / collected amount × cpm in filter paper after washing = total antibody-bound radioactivity * the amount of isotope bound to the antibody should be in the sample 70-95% of the total isotope. Key points and description of the experiment Iodination by the chloramine T method can also be carried out in a buffer solution of about pH 7.0. Both must ensure that no reducing agent is present in the reaction system. If the oxidation reaction damages the protein, the amount of chlorinated antibody and fluorescein isothiocyanate (FITC) amine T can be reduced to 0.02mg / ml, and the concentration of the heavy sulfurous acid solution can be reduced to 0.024mg / ml. Iodogen-coated test tubes can be stored in a desiccator at room temperature for many years. The 125I-labeled antibody can be used within six weeks after preparation (the half-life of 125I is 59.6 days). Care should be taken to ensure that the Na125I used is fresh and the specific activity of the old formulation is low. Tyrosine iodination occasionally interferes with the antibody binding site of the antigen, thus reducing its binding capacity, but this is rare. * The above isotope labeling method requires highly purified antibodies to ensure reliable results. References Fraker, PL .and Speck, JC (1978) Protein and cell membrane iodination with sparkingly soluble chloramines, 1,3,4,6-tetrachloro-3α, 6α-diphenyl-glucoluril. Biochem. Biophys. Res. Commun. 80,849 -857. Greenwood, FC., Hunter, WM. And Glover, JS. (1963) The preparation of 125I-labeled human growth hormone of high specific radioactivity. J. Biochem. 89, 114-123. Sun Li, Zhu Zhengmei and Gu Tianjue radiolabeled lectin "Sugar Complex Biochemical Research Technology" Zhejiang University Press 1999 pp.501-504 (Zhu Zhengmei) Basic Principle of Antibody Enzyme Labeling Method This method uses a coupling agent to bind the enzyme to the antibody. That is, through the application of single, double or multifunctional reagents, respectively react with the functional groups present in the antibody of the macromolecule to generate an enzyme-antibody coupling complex. Among the different preparation methods of enzyme-antibody complexes, the most widely used method is the method of adding glutaraldehyde in the first step. Compared with other coupling methods, this method has the advantages of simple operation, mild reaction conditions, and wide practicality. . Reagents and instruments Affinity-purified polyclonal antibodies or biochemically pure monoclonal antibodies (5 mg / ml PBS solution or 0.1 mol / L phosphate buffer pH 6.8) enzymes used for labeling (EIA grade, commercially available): Horseradish peroxidase (HRP, purity A430 / A275> 3), Aspergillus niger glucose oxidase, calf intestinal alkaline phosphatase (AKP), or Escherichia coli β-D-galactosidase can be used , But HRP is most commonly used. 25% glutaraldehyde solution (electron microscope grade) 0.1 mol / L potassium phosphate buffer (pH6.8) PBS (see appendix) 2 mol / L glycine solution steamed glycerin dialysis bag (molecular weight 6000-8000) electromagnetic stirrer and stir bar Operation step 1. Antibody and peroxidase coupling 5mg antibody and 10mg enzyme in a total volume of 1ml of 0.1 mol / L potassium phosphate buffer (pH 6.8) mixed; at 4 ℃ 0.1 mol / L potassium phosphate buffer (pH6.8) dialysis overnight; dilute glutaraldehyde to 1% with 0.1 mol / L potassium phosphate buffer (pH6.8); add 50 μl of diluted glutaraldehyde to the dialysis mixture, lightly 1 at room temperature Stir for three hours; add 2 mol / L glycine solution to make the final concentration reach 0.1 mol / L, leave the mixture at room temperature for 2 hours, and block the remaining aldehyde-based mixture to dialyze against PBS overnight at 4 ° C; centrifuge at 10000 × g at 4 ° C for 30 hours Divide; transfer the supernatant to another tube, add glycerol according to the volume of 1: 1, so that the final concentration reaches 50%; store at -20 ℃.
2. Antibody and AKP coupling Mix 5mg antibody and 10mg enzyme in a total volume of 2ml of 0.1mol / L potassium phosphate buffer (pH 6.8); other operations are the same as HRP labeling method.
3. Coupling of antibody and glucose oxidase According to the HRP coupling method, only 1% glutaraldehyde added was changed to 150 μl. 4. The antibody is coupled with β-D-galactosidase according to the HRP coupling method, but the total volume of the coupling reaction is changed to 2ml, and the amount of 1% glutaraldehyde is changed to 100μl. The quality of the reaction can be measured by direct enzyme immunoassay using a microplate coated with antigen. (For specific methods, please refer to -----) The main points and instructions of the experiment. If the amino group involved in the coupling is located at the binding site between the antibody and the antigen, the affinity of the labeled antibody will be destroyed to varying degrees during the coupling reaction; The main reason for affecting the success rate of coupling is that there is free amino group in the reaction mixture, and the free amino group easily reacts with glutaraldehyde, thus interfering with protein cross-linking. Therefore, the buffer must be prepared with water that is absolutely free of organic matter, and the reaction mixture must be fully dialyzed against the buffer before coupling, which is the key to successful reaction. Tris-glycine buffer cannot be used for the reaction.
References Avrameas, S. (1969) Coupling of enzymes to protein with glutaraldehyde. Use of the conjugates for the detection of antigens and antibody. Immunochemistry 5, 43-52 Avrameas, S. and Ternyck, T. (1971) Peroxidase labelled antibody and Fab congugates with enhanced intracellular penetration. Immunochemistry. 8 (12): 1175-1179 (Zhu Zhengmei)
Section 3 Basic Principles of Antibody Biotinylated Labeling This method allows the ε-amino group of antibodies or other proteins to be covalently bound to the acylated biotin through the arm. Thereafter, biotinylated molecules can be detected using enzyme label-avidin or fluorescent dye-streptavidin complexes. The small molecule water-soluble biotin has a high affinity for the bacterial protein streptavidin is the design basis of this method. Reagents and instruments NHSB: N-hydroxysuccinimide biotin (commercially available) 0.1 mol / L sodium bicarbonate buffer, pH 8.0 (see appendix) Double distilled water (note the removal of free amino groups) to prepare bicarbonate Sodium buffer or boric acid buffer DMSO (dimethyl sulfoxide, toxic reagent) 1 mol / L NH4Cl sodium azide (toxic reagent) PBS (see appendix) 10g / L bovine serum albumin (W / W) PBS liquid molecular sieve column (E.g. G25) Electromagnetic stirrer dialysis bag (MW CO 8000) Aluminium platinum micropipette operation steps The protein to be biotinylated is buffered with 0.1 mol / L sodium bicarbonate buffer (pH 8.0) or 0.5 mol / L boric acid The solution (pH 8.6) is diluted to 1 mg / ml, and the biotinylated volume for general laboratory applications is 1-2.5 ml; alternately use 0.1 mol / L sodium bicarbonate buffer (pH 8.0) or 0.5 mol / L boric acid buffer ( pH 8.6), thoroughly dialyze the protein; dissolve 1 mg of NHSB with 1 ml of DMSO; add 120 μl of NHSB solution (ie containing 120 μg of NHSB) to 1 ml of protein solution (ie containing 1 mg of protein); keep stirring at room temperature and keep warm for 2-4 hours; Add 9.6 μL of 1 mol / L NH4Cl (add 1 μl per 25 μg NHSB) and stir at room temperature for 10 minutes; at 4 ° C, dialyze PBS thoroughly to remove free biotin; 1ml of product on a molecular sieve column, eluting with PBS slowly, collecting 1ml / tube, between the protein wash 1-3ml; Finally, the sample was added sodium azide (final concentration of 0.5g / L) and 1.0g / L BSA. Store the combined product at 4 ℃, protected from light, or add 50% redistilled glycerin, and store at -20 ℃. Quality monitoring Avidin, streptavidin or neutravidin combined with enzyme- or fluorescent dye-, the cross-linking rate is determined by standard methods (Western blotting or immunofluorescence staining). Key points of the experiment and explanations If there is sodium azide or free amino groups in the reaction mixture, the labeling reaction will be inhibited. Therefore, the protein should be fully dialyzed against 0.1 mol / L sodium bicarbonate buffer or 0.5 mol / L boric acid buffer before the reaction; the molecular ratio between the NHSB used and the protein to be biotinylated is based on the ε-amino group of the protein surface The density will be different. Improper selection will affect the efficiency of the label. Several different molecular ratios should be used to screen the optimal conditions first. Excessive use of NHSB is also disadvantageous. The antigen binding site may be blocked, resulting in antibody loss. Live; because the amino group of the antibody is not easily accessible, it may cause insufficient biotinylation. At this time, detergents such as Triton X-100, Tween 20, etc. can be added. When the free epsilon-amino group (the amino group of the lysine residue) is present at the antigen binding site of the antibody or at the catalytic site of the enzyme, biotinylation reduces or impairs the binding capacity or activity of the antibody protein. At this time, other cross-linking methods should be tried; biotin may also be combined with different functional groups, such as carbonyl, amino, mercapto, isoimidazolyl, and phenol groups, or covalently bonded to sugar groups (see Reference 1 for details) ; After the cross-linking reaction, it should be fully dialyzed, otherwise, the residual biotin will have a competitive effect on the binding of biotinylated antibody and avidin; In the fluorescent labeling experiment of cells, the background of neutralizing avidin is low, but Because streptavidin contains a small amount of positive charge, it can cause a high background for some cells.
References Bayer, EA. And Wilchek, M. (1980) The use of avidin-biotin complex as a tool in molecular biology. Meth. Biochem. Anal. 26, 1-45. Guesdon, JL., Ternynck, T. And Avrameas, S. (1979) Thr use of avidin-biotin interaction of immuenzymatic techniques. J. Histochem. Cytochem. 27, 1131-1139. (æœ±æ£ ç¾Ž)
Section 4 Basic Principles of Antibody Fluorescent Labeling Many protein molecules contain more lysine residues on their surface. The free ε-amino groups of these lysine residues can be covalently bound to FITC (the excitation wavelength is 492 nm and the emission wavelength is 525 nm). Antibodies that bind to FITC can be used as specific probes to determine the presence of corresponding antigens in cells. FITC has a very high quantum yield (ratio of emitted light to absorbed light, 0.85), and the stability of the formed conjugate is very good. FITC is the most widely used fluorescent dye. According to the characteristics of FITC, the flow cytometer has designed a laser wavelength of 488 nm, which is very close to FITC's maximum excitation wavelength of 492 nm). The coupling reaction is a nucleophilic reaction between the free ε-amino group of the lysine residue and FITC at pH 9.8, thereby forming a thiourea linkage. Reagents and instruments to be coupled antibody sodium bicarbonate buffer: 25m mol / L Na2CO3 / NaHCO3 buffer pH9.8 (freshly prepared, see appendix for the preparation method) PBS (see appendix) sodium azide (toxic reagent) isothiocyanate Fluorescein (I type heterologous, commercially available) Electromagnetic stirrer Aluminum platinum operation steps Dilute the antibody to 1-5 mg / ml with sodium bicarbonate buffer pH 9.8, or the antibody is dialyzed sufficiently against the buffer to Lysine does not dissociate (remove its positive charge), but pay attention to keep most of the protein still not denatured; place the dialysis bag in 100ml of pH9.8 sodium bicarbonate buffer containing 0.1 mg / ml FITC (new preparation) In the beaker, wrap the beaker with aluminum platinum to protect from light and stir overnight at 4 ° C; the above antibody solution was dialyzed against PBS at 4 ° C to stop the reaction. In the meantime, change the PBS solution at least three times until the absorption at 480nm is zero; add 0.5g / L sodium azide. After that, the conjugate was stored at 4 ° C in the dark or frozen at -20 ° C after aliquoting. The detection of the quality of the fluorescence coupling uses a standard immunofluorescence staining test to determine the coupling rate. Or use F / P measurement to identify the quality of its FITC label: Take 0.2ml of the conjugate solution, add 2.8ml of PBS (or both use half amount), measure its A490 / A280, check the FITC mark curve to get its concentration μg / ml value, according to the extinction coefficient of IgG (mg / ml about A280 1.2), the F / P value can be calculated, that is, the ratio of μgFITC labeled per mg antibody, which can identify and compare the quality of each batch of markers. Key points of the experiment and explanation The coupling reaction needs to be carried out in a solution as close to pH 9.8 as possible, and pay attention to maintaining this pH level during the reaction; make sure that the coupling reaction buffer does not contain free amino groups (Tris, ammonia and Sodium azide can react with FITC, so it will reduce the coupling rate of protein and FITC); the ratio of FITC to protein (F / P) can be identified by measuring the absorbance at 495nm and 280nm. The range of this ratio should be 0.3-1.0 (see the method above); the only limitation of FITC is that it is easily quenched by light, so the complex must always be protected from light; if the FITC-complex is not sufficiently dialyzed against PBS, it may cause high In the background, it interferes with immunofluorescence staining; if the labeled protein is a secondary antibody or a universal primary antibody, it may be more convenient and desirable to purchase from a commercial product.
References The, TH and Feltkamp, ​​TEW. (1970) Conjugation of f luorescein isothiocyanate to antobodies. I. Experiments on the conditions of conjugation. Immunology 18, 865-873. The, TH and Feltkamp, ​​TEW. (1970) Conjugation of fluorescein isothiocyanate to antobodies. II. A reproducible method. Immunology 18, 875-881 (æœ±æ£ ç¾Ž)
Section 5 Basic Principles of Biosynthesis Process Marking The biosynthesis process is marked mainly to study the biochemical properties, synthesis, processing, intracellular transmission, secretion and degradation processes of proteins. The cells are usually placed in a medium containing sufficient nutrients and radiolabeled amino acids. Although the content of methionine and cysteine ​​in protein is low, the 35S marker has high specificity (> 2.93 × 1013 Bq / mmol ), Easy to detect, so it is the first choice amino acid for biosynthesis labeling. The following steps are methods for short-term labeling of cells (single cell suspension or culture of spleen or thymus) in suspension. Note: The laboratory should be equipped with various related equipment for radioactive compound operations. 1. Special water bath, incubator and centrifuge for radioactive work; 2. During the labeling operation, use a Geiger counter to monitor the operation area; 3. Do the 35S solid and liquid waste disposal work; 4. In the experiment Before you start, you must obtain the approval of the relevant departments; in operation, you must strictly comply with the requirements of the national management regulations. Reagents and equipment Cells in culture medium ice-cooled PBS (see Appendix A) labeled medium: RPMI 1640 medium without methionine and cysteine, preheated to 37 ℃ in water bath [35S] methionine and [35S] Cysteine ​​(800-l200 Ci / mmol = 2.93-4.44 × 1013 Bq / mmol) 15 or 50 ml conical polypropylene centrifuge tube to fully dialyze the labeled medium with fetal bovine serum (FCS) L-glutamic acid. Micropipette incubator centrifuge (freezing) operation steps (1) Preparation of cells At room temperature, centrifuge at 300 × g for 5 minutes to collect 107-108 cells; in a conical centrifuge tube, use 10ml of 37 ℃ Wash the cells with labeling medium, then centrifuge at 300 × g for 5 minutes; discard the supernatant; add the cell pellet to the labeling medium and repeat washing once. (2) Pre-pulse washed cells, resuspended in 37 ℃ preheated labeled medium (4 ml containing 20 × 106 cells), incubated at 37 ℃ for 30 minutes, intermittent vortex to deplete the original cells Methionine and Cysteine; Thaw at room temperature [35S] Methionine and [35S] Cysteine; Note: [35S] Methionine is volatile, please open [35S in a dedicated fume hood equipped with activated carbon filter ] Methionine stock solution. Currently, non-volatile [35S] methionine and [35S] cysteine ​​are both commercially available. The cell solution was centrifuged at 300 × g for 5 minutes at room temperature, and the supernatant was discarded. (3) Pulse Pulse resuspends the cell mass in labeled medium (concentration 20 × 106 cells / 1 ml); add 250 μCi of [35S] methionine and [35S] cysteine ​​(9.25 × 109 Bq); Put the cells in a 37 ℃ incubator and incubate for 30 minutes to 3 hours. During the incubation, shake frequently to keep the cells suspended; the cell solution is centrifuged at 300 × g for 5 minutes at room temperature, and the supernatant is discarded; Warning: used culture Both the base and the washing solution contain radioactivity, so please operate and dispose of the radioactive material according to the prescribed method; resuspend the cells with 10ml of ice-cold PBS and wash twice; according to the method in "Preparation of Cell Extraction" (see Protocol 43) Perform cell processing and analysis, or freeze cells at -20 ℃. Quality points If the pulse needs to last more than 1 hour, it is necessary to add 2% fetal bovine serum and 2% L-glutamic acid to the labeling medium under sterile conditions; for most types of cells, you can use Screw-tissue tissue culture flask, and incubate in a humidified 5% CO2 incubator; incubation must be carried out at 37 ℃, low temperature will significantly reduce the radioactivity incorporated into the protein; 14C-labeling can also be used Amino acids are labeled, and their half-life is long, which is conducive to the use of labeled proteins for a long time (several weeks), such as the monoclonal antibodies secreted by hybridoma B cells, often using 14C-labeled amino acid mixtures (serine , Leucine, Valine).
References Coligan, JE, Gates, FT, III, Kimball, ES and Maloy, WL (1983) Radiochemical sequence analysis of biosynthetically labeled proteins. Meth. Enzymol. 91,413-434. Meisenhelder, J. and Hunter, T. (1988) Radioactive protein labelling techniques. Nature (London) 335, 120. (Li Jinghua Zhu Zhengmei)
Section VI Basic Principles of Immunocolloid Gold-labeled Antibodies and Detection Antigens Colloidal gold is an effective marker in light microscope and electron microscope, and can detect single and multiple antigens. Colloidal gold is unstable in electrolytes, but colloidal gold coated with protein is stable. Generally, immunoglobulin or protein A is used to coat colloidal gold, which can be used as a labeled secondary antibody for immunodetection. This method has a wide range of applications and the stained samples can be stored for a long time. Reagents and equipment Ultracentrifuge microscope incubator spectrophotometer 0.2 um microporous filter glass slide, cover sheet, filter paper sodium chloraurate 1% sodium citrate aqueous solution goat anti-mouse Ig antibody specific mouse anti-human to T lymphocytes Monoclonal antibody isolated from human peripheral blood leukocyte suspension 10% sodium chloride 1% polyethylene glycol 0.01mol / L pH7.2 PBS 1% glutaraldehyde ethanol solution 50% silver nitrate solution (prepared one day in advance) gelatin development Solution, 2g of gelatin is dissolved in 99ml of distilled water, add 1ml of formic acid operation steps Preparation of colloidal gold solution Weigh 0.1g of sodium chloroaurate and dissolve in 1L of deionized water; heat to boil, add vigorously and quickly add 25ml % Sodium citrate solution; continue to boil for 5 minutes until the solution turns orange-red; adjust the optical density absorption value of the solution at 525 nm wavelength to 0.8 with distilled water. Coating with colloidal gold The goat anti-mouse Ig antibody was centrifuged at 36 900 g for 30 minutes; the supernatant was filtered through a 0.2 nm filter; the optimal concentration of protein coating was determined: the protein was serially diluted 10-fold each 1ml, and 5ml was added For the colloidal gold solution, add 1 ml of 10% sodium chloride solution after 1 minute and let it rest for 5 minutes; use the colloidal gold solution as a blank control to measure the light absorbance, and select the protein concentration with the lowest absorption value for formal coating. Take 50ml of colloidal gold solution, adjust the pH value to 7.6 with 0.2M K2CO3, add the protein according to the determined optimal ratio and quickly mix, after the protein is adsorbed for 2 minutes, add 0.5ml 1% polyethylene glycol to prevent non-specific aggregation ; Centrifuge at 5000g for 1 hour, discard the supernatant, suspend the colloidal gold in PBS containing 1% polyethylene glycol; repeat once; discard the supernatant, and coat the colloidal gold solution with 5 ml containing 1% BSA Diluted in PBS, filtered through a microporous filter, and stored at 4 ℃. (3) Antigen detection Take 20ul of human peripheral blood leukocytes and 5l T lymphocyte-specific monoclonal antibody and incubate at 4 ° C for 30 minutes; 1000 r / separation heart washing cells for 5 minutes, 2 times; the washed cells and 20l ( General titer 1:20) Colloidal gold-labeled goat anti-mouse Ig antibody was incubated at room temperature for 30-60 minutes; 1000 r / separation heart washes the cells for 5 minutes, 2 times; smear the cells on a glass slide with 1% Fix the cells with glutaraldehyde for 10 minutes and wash the excess fixative; place the slide cells upside down in a Petri dish with moistened filter paper, add 4 drops of 50% silver nitrate solution and 2 drops of gelatin coloring solution, and cover with Put the slides in a 60-65 ° C incubator and develop the color for 3-4 minutes until the slide specimens appear golden brown; remove the coverslips, rinse them quickly with distilled water for a few seconds, and dry them; observe under an optical microscope. For the control experiment, cells without primary antibody can be used as the control experiment group. The main points and instructions of the test should be very clean, need to be silanized; use high-purity distilled deionized water multiple times; the formation, size and speed of colloidal gold particles all determine the stability of colloidal solution, water quality and glass The surface is very important for starting reduction and stabilizing the colloid. The diameter of the colloidal gold particles prepared in this process is about 18-20 nm; because the colloidal gold is unstable in the electrolyte, the entire operation process of this experiment should be strict and rapid, pay attention to the liquid color; protein package The relative optimal concentration of the colloidal gold should be determined in advance. When determining the optimal concentration, the pH of the colloidal gold solution should also be adjusted to pH 7.6; the dilution concentration of the primary antibody and the colloidal gold labeled secondary antibody should be pre-tested in advance The best titer. References Yang Hanmin (1997) "Cell Biology Experiments" Second Edition Higher Education Press Chief Editor Yang Jingshan (1990) "Medical Cell Chemistry and Cell Biotechnology" Beijing Medical University and China Union Medical University United Press (Ge Changhui)
This is a NEW kind of professional USB Flash Disk Printer, its productivity equivalent to R1390 printer and can realize automatic continuous printing .Usb Flash Disk Printer can be inserted adapter, computer –controlled, continuously print out 60pcs discs at once. Cyclic loading can also printing item, to achieve uninterrupted printing.
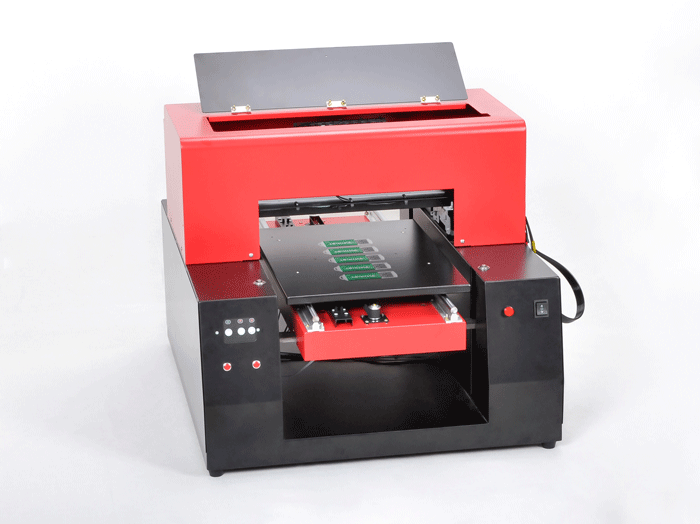
USB Flash Disk Printer
Usb Flash Disk Printer,Printer For Usb Flash Disk,Usb Flash Disk Logo Printer,Usb Flash Disk Shell Printer,Two Color Usb Flash Disk Printer
Shenzhen Refinecolor Technology Co., LTD. , https://www.rfcprinter.com